Scientific objectives of MTH 2.0
The MetaboHUB infrastructure workplan has been successfully evaluated by the ANR international panel and will be prolonged over the 2020-2025 period to tackle these challenges.
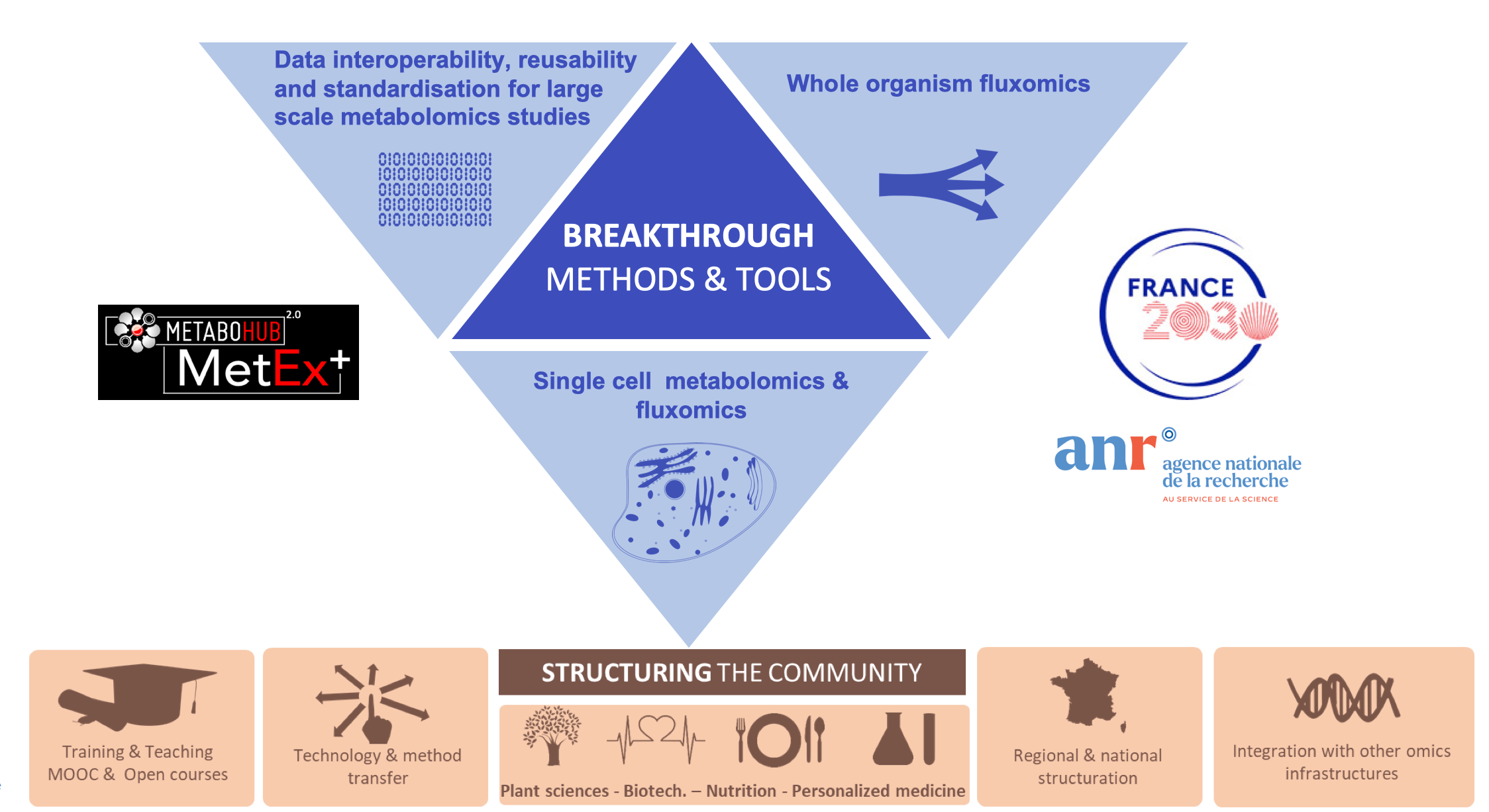
In this context, MTH partners have identified three challenges to tackle:
- Data interoperability, reusability and standardisation for large scale metabolomics studies
- Single cell metabolomics and fluxomics
- Whole body fluxomics
The MetaboHUB 2.0 Equipex plus project
To implement this project, MTH 2.0 defined a scientific strategy focused on key methodological and technological research activities. This roadmap will allow to keep the leadership in metabolomics and fluxomics and to serve many life science communities.
(i) Advanced methods for comprehensive metabolomics for large cohort studies. The aim is to develop and implement new high-throughput multi-event and quantitative data acquisition methods, data processing workflows, as well as advanced methods for knowledge extraction. Standard operating procedures will be disseminated to the scientific community. This should foster data standardization, reusability and interoperability.
(ii) Extending metabolite identification and annotation of metabolomics datasets. We will foster the combined use of MSn experiments and ion mobility spectroscopy using cutting-edge instruments to characterize unknown mass spectrometry features present in biological matrices of interest. Special attention will be placed towards complex metabolites (e.g. plant specialised compounds, toxicants) and lipids. These data will be put into the MetaboHUB spectral database (PeakForest). New algorithms for automated processing and analysis of such data will have to be developed and implemented.
(iii) Microfluidic tools for in situ and single-cell metabolomics and fluxomics. Sample preparation will be miniaturized thanks to microfluidics. This will enable to handling samples with a limited number of cells (i.e., 1000 to 100 cells), or to sorting specific cells or cell types for further MS-based metabolomics and fluxomics analysis. A specific objective is to develop single cell investigations to highlight and understand metabolic differences existing between seemingly homogenous populations of cells. Microfluidic tools should also enable the detection of metabolites present at (ultra)trace levels in relevant biological matrices.
(iv) Bedside metabolism: clinical fluxomics. The ambition of MetaboHUB 2.0 is to set-up an innovative clinical fluxomics infrastructure, unique in France and in Europe, enabling to directly capture the true complexity of whole-body metabolism and its disturbances in patients, for paving the way to the next-generation precision medicine and nutrition. All relevant installations, workflows, data management & processing tools, and mathematical frameworks will be developed for being able to investigate metabolism at the whole organism level from in vivo (stable) isotope tracing experiments. This will be achieved in collaboration with the Clinical Investigation Center at Toulouse Hospitals (Pr. P. Gourdy) where all relevant clinical set-ups for infusion experiments (of non-labelled substrates) in Human and e.g. clamp experiments are already established in compliance with all ethical regulations. The major applications will be precision diagnosis, drug targeting & validation, personalized medicine, in the fields of cancer, metabolic diseases, and aging.
- (v) Creating FAIR methods and tools for knowledge mining. FAIR, among which interoperability between data and tools, is an acknowledged challenge in metabolomics and fluxomics due to the variety of analytical platforms and sample metadata. The aim will be to unlock these computational challenges together with the French Bioinformatics Institute (IFB).
Analysis of equipment needs
Achieving these research activities requires three kinds of new instruments, which are the subject of the MetaboHUB 2.0 Equipex plus project :
- 6 new generation ultra-high resolution mass spectrometers with ultra-high performance chromatography systems enabling sensitive metabolite detection and ultra-fast advanced annotation possibilities that help the identification and quantification for increased phenotyping capabilities
- Microfluidic devices (a multifunction laser photolithography system and a 3D laser printer) for the production of microfluidic chips to be coupled to MS and NMR instruments to improve sensitivity and deal with small amount of biological materials and
- NMR microfluidic probe for low volume analysis compatible with microfluidic systems for sensivity improvement. By significantly strengthening the potential of MTH platforms, these equipments should make it possible to accelerate the pace of discoveries and innovations stemming from living science research and to develop synergies with economic players.
A unique setup in France and in Europe serving a wide range of scientific communities
These technological research activities will be developed around selected scientific projects in collaboration with both partner units and external strategic actors in the fields of plant sciences, biotechnology, human nutrition and personalized medicine (cancer, metabolic diseases, aging, gastroenterology and Hepatology). To this end, we will develop and extend collaborations with other national infrastructures such as Profi and IFB, with industrial demonstrators such as Metagenopolis, TWB and IBISBA, and with other EQUIPEX plus projects involved in translational medicine.
Finally, MetaboHub2.0 aims at creating and making available for the scientific community a unique setup in France and in Europe with the potential to create methods and tools which could not be developed without the synergetic efforts of its 5 regional facilities. By tackling these current methodological and technological bottlenecks, MetaboHub2.0 will provide the scientific community with analytical and computational solutions to go beyond current knowledge on metabolism. Since 2013, MetaboHub has shown its ability to transfer its technological and innovative developments to research laboratories and facilities and will persue its community-structuring activity through its tight connection with the national metabolomics and fluxomics society (RFMF) and by leading the regional structuration of the field. With the implementation of newly integrated services, the MetaboHUB2.0 EQUIPEX plus project will foster the development of a more interoperable metabolomics, which will accelerate its translation into the various application fields.